Pharmokinetics Modelling Project
Overview
Teaching: 30 min
Exercises: 0 minQuestions
What is the project you will be working on?
How will the group projects work?
Objectives
Explain the background and the project you will be doing for the next 3 days.
How each group will work together.
Assessment deliverables
Pharmokinetic Modelling
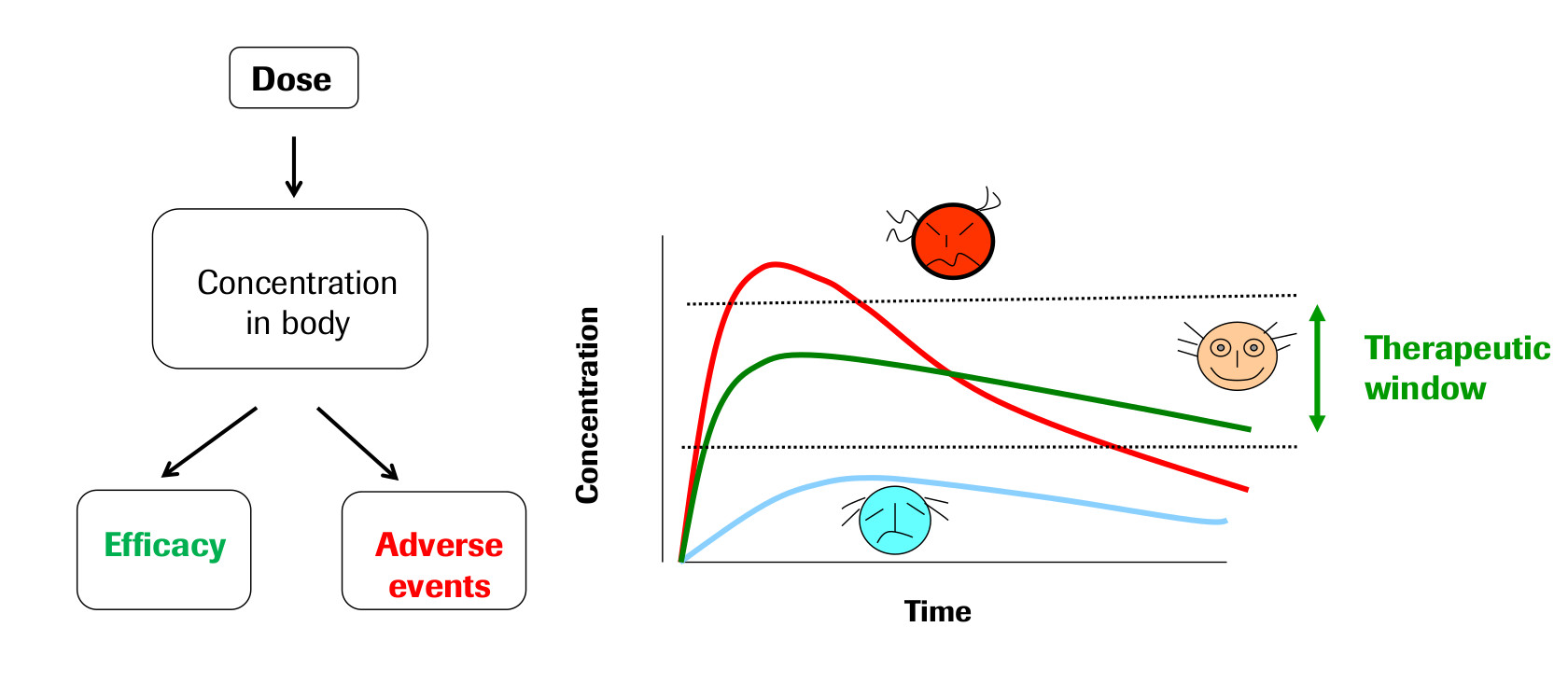
The field of Pharmacokinetics (PK) provides a quantitative basis for describing the delivery of a drug to a patient, the diffusion of that drug through the plasma/body tissue, and the subsequent clearance of the drug from the patient’s system. PK is used to ensure that there is sufficient concentration of the drug to maintain the required efficacy of the drug, while ensuring that the concentration levels remain below the toxic threshold (See Fig 1). Pharmacokinetic (PK) models are often combined with Pharmacodynamic (PD) models, which model the positive effects of the drug, such as the binding of a drug to the biological target, and/or undesirable side effects, to form a full PKPD model of the drug-body interaction. This project will only focus on PK, neglecting the interaction with a PD model.
PK enables the following processes to be quantified:
- Absorption
- Distribution
- Metabolism
- Excretion
These are often referred to as ADME, and taken together describe the drug concentration in the body when medicine is prescribed. These ADME processes are typically described by zeroth-order or first-order rate reactions modelling the dynamics of the quantity of drug $q$, with a given rate parameter $k$, for example:
\[\frac{dq}{dt} = -k^*,\] \[\frac{dq}{dt} = -k q.\]The body itself is modelled as one or more compartments, each of which is defined as a kinetically homogeneous unit (these compartments do not relate to specific organs in the body, unlike Physiologically based pharmacokinetic, PBPK, modeling). There is typically a main central compartment into which the drug is administered and from which the drug is excreted from the body, combined with zero or more peripheral compartments to which the drug can be distributed to/from the central compartment (See Fig 2). Each peripheral compartment is only connected to the central compartment.
The following example PK model describes the two-compartment model shown diagrammatically in Fig 2. The time-dependent variables to be solved are the drug quantity in the central and peripheral compartments, $q_c$ and $q_{p1}$ (units: [ng]) respectively.
\[\frac{dq_c}{dt} = \text{Dose}(t) - \frac{q_c}{V_c} CL - Q_{p1} \left ( \frac{q_c}{V_c} - \frac{q_{p1}}{V_{p1}} \right ),\] \[\frac{dq_{p1}}{dt} = Q_{p1} \left ( \frac{q_c}{V_c} - \frac{q_{p1}}{V_{p1}} \right ).\]This model describes an intravenous bolus dosing protocol, with a linear clearance from the central compartment (non-linear clearance processes are also possible, but not considered here). The input parameters to the model are:
- The dose function $\text{Dose}(t)$, which could consist of instantaneous doses of $X$ ng of the drug at one or more time points, or a steady application of $X$ ng per hour over a given time period, or some combination.
- $V_c$ [mL], the volume of the central compartment
- $V_{p1}$ [mL], the volume of the first peripheral compartment
- $V_{p1}$ [mL], the volume of the peripheral compartment 1
- $CL$ [mL/h], the clearance/elimination rate from the central compartment
- $Q_{p1}$ [mL/h], the transition rate between central compartment and peripheral compartment 1
Another example model we will show uses subcutaneous dosing, and adds an additional compartment from which the drug is absorbed to the central compartment:
\[\frac{dq_0}{dt} = \text{Dose}(t) - k_a q_0,\] \[\frac{dq_c}{dt} = k_a q_0 - \frac{q_c}{V_c} CL - Q_{p1} \left ( \frac{q_c}{V_c} - \frac{q_{p1}}{V_{p1}} \right ),\] \[\frac{dq_{p1}}{dt} = Q_{p1} \left ( \frac{q_c}{V_c} - \frac{q_{p1}}{V_{p1}} \right ),\]where $k_a$ [/h] is the “absorption” rate for the s.c dosing.
Project Description
Your group project is to design and implement a python library that can specify, solve and visualise the solution of a PK model. You might find it helpful to base this on a “starter” repository which you can clone from https://github.com/sabs-r3/software-engineering-projects-pk. This contains a basic Python script that constructs a two-compartment PK model with i.v. dosing, solves it using the Scipy
solve_ivpODE solver, and then plots the results usingMatplotlib. It also contains the framework of a basic python package that can be installed usingpip. This package uses theunittesttesting framework, and defines a few classesModel,Protocol, andSolution. Note that you can feel free to use as much or as little of this template repository as you like, it is a suggestion based on a specific set of design decisions, and many other choices could be equal or superior to those chosen in the provided template. Your PK modelling library should ideally have the following functionality:
- pip installable
- github repository, with issues + PRs that fully document the development process
- unit testing with a good test coverage
- fully documented, e.g. README, API documentation, OS license
- continuous integration (e.g. Github actions) for automatic testing and documentation generation
- The ability to specify the form of the PK model, including the number of peripheral compartments, the type of dosing (intravenous bolus versus subcutaneous), and the dosing protocol.
- Users can specify the protocol independently from the model (e.g. be able to solve a one and two compartment model for the same dosing protocol)
- Ability to solve for the drug quantity in each compartment over time, given a model and a protocol
- Ability to visualise the solution of a model, and to compare two different solutions.
- Something else? Feel free to suggest alternative features.
Numerical solution of ODEs using Scipy
In the starter template, Scipy’s
solve_ivpis used to solve the set of ODEs that define the PK model. Since we are using linear PK, we could have used analytical solutions, but relying on an ODE solver allows us to be more flexible if we wish to include non-linear models in the future. In addition, you will need to be able to numerically solve ODE models for the Mathematical Modelling module in weeks 6-7, so take this time to become familiar with how to usesolve_ivp.
Structure of the project days
Each project team is free to organise their interaction according to what works best for the group. You are free to meet as a group in the DTC, and each group will be assigned a student room in the building. Each group will also have a breakout room to meet virtually if required. Each group should create issues on their repository corresponding to individual work items, and assign these members of the group. Each group should identify a workflow that they will use to add code to the repository (e.g. one branch per issue, then a pull request once the work is ready to add to the main branch).
Hand-in
Nothing needs to be physically handed-in for this assessment other than a URL for the GitHub repository for each group. Your feedback and assessment for the software engineering module will be based on this repository: the source code contained within, the generated documentation, and the content of the issues and pull requests. The source code should conform to a consistent style throughout, and docstrings & comments should be appropriate so that the code can be easily read and understood. The documentation should be sufficient so that a new user could easily use the library. The issues and pull requests should be sufficiently detailed so that a developer could join the project half-way through and get up-to-speed on the current state of development and what needs to be done.
Key Points
The groups will be identical to the SABS year-long open source software groups.
Project management will consist of (at least) daily stand-ups and the use of GitHub issues and pull requests.
Submit your GitHub repository for assessment by 5:30pm Friday 22nd October.