Diagnosing Issues and Improving Robustness
Overview
Teaching: 30 min
Exercises: 15 minQuestions
Once we know our program has errors, how can we identify where they are?
How can we make our programs more resilient to failure?
Objectives
Use a debugger to explore behaviour of a running program
Describe and identify edge and corner test cases and explain why they are important
Apply error handling and defensive programming techniques to improve robustness of a program
Finding faults in software
Unit testing can tell us something’s wrong and give a rough idea of where the error is by what test(s) are failing. But it doesn’t tell us exactly where the problem is (i.e. what line), or how it came about. We can do things like output program state at various points, perhaps using print statements to output the contents of variables, maybe even use a logging capability to output the state of everything as the program progresses, or look at intermediately generated files to give us an idea of what went wrong.
But such approaches only go so far and often these are time consuming and aren’t enough. In complex programs like simulation codes, sometimes we need to get inside the code as it’s running and explore. This is where using a debugger can be useful.
Normalising patient data
We wish to add a new function to our inflammation example, one that will normalise a given inflammation data array so that all the entries lie between 0 and 1.
Add a new function to inflammation/models.py called patient_normalise(), and copy
the following code:
def patient_normalise(data):
"""Normalise patient data between 0 and 1 of a 2D inflammation data array."""
max_for_each_patient = np.max(data, axis=0)
return data / max_for_each_patient[:, np.newaxis]
So here we’re attempting to normalise each patient’s inflammation data by the maximum inflammation experienced by that patient, so that the final values are between 0 and 1. We find the maximum value for a patient, and using NumPy’s elementwise division, divide each value by that maximum. In order to prevent an unwanted feature of NumPy called broadcasting, we need to add a blank axis to our array of patient maximums. Note there is also an assumption in this calculation that the minimum value we want is always zero. This is a sensible assumption for this particular application, since the zero value is a special case indicating that a patient experiences no inflammation on that day.
Now add a new test in tests/test_models.py, to check that the normalisation function
is correct for some test data.
@pytest.mark.parametrize(
"test, expected",
[
([[1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0.33, 0.66, 1], [0.66, 0.83, 1], [0.77, 0.88, 1]])
])
def test_patient_normalise(test, expected):
"""Test normalisation works for arrays of one and positive integers."""
from inflammation.models import patient_normalise
npt.assert_almost_equal(np.array(expected), patient_normalise(np.array(test)), decimal=2)
Note the assumption here that a test accuracy of two decimal places is sufficient!
Run the tests again using pytest tests/test_model.py and you will note that the new
test is failing, with an error message that doesn’t give many clues as to what is wrong
E AssertionError:
E Arrays are not almost equal to 2 decimals
E
E Mismatched elements: 6 / 9 (66.7%)
E Max absolute difference: 0.57142857
E Max relative difference: 1.33333333
E x: array([[0.33, 0.66, 1. ],
E [0.66, 0.83, 1. ],
E [0.77, 0.88, 1. ]])
E y: array([[0.14, 0.29, 0.43],
E [0.5 , 0.62, 0.75],
E [0.78, 0.89, 1. ]])
tests/test_models.py:53: AssertionError
Pytest and debugging in VSCode
Let’s use a debugger to see what’s going on and why the function failed. Think of it like performing exploratory surgery - on code! Debuggers allow us to peer at the internal workings of a program, such as variables and other state, as it performs its functions.
First we will set up VSCode to run and debug our tests. If you haven’t done so already,
you will first need to enable the PyTest framework in VSCode. You can do this by
selecting the Python: Configure Tests command in the Command Palette (Ctrl+Shift+P).
This will then prompt you to select a test framework (Pytest), and a directory
containing the tests (tests). You should then see the Test view, shown as a beaker, in
the left hand activity sidebar. Select this and you should see the list of tests, along
with our new test test_patient_normalise. If you select this test you should see some
icons to the right that either run, debug or open the test_patient_normalise test. You
can see what this looks like in the screenshot below.
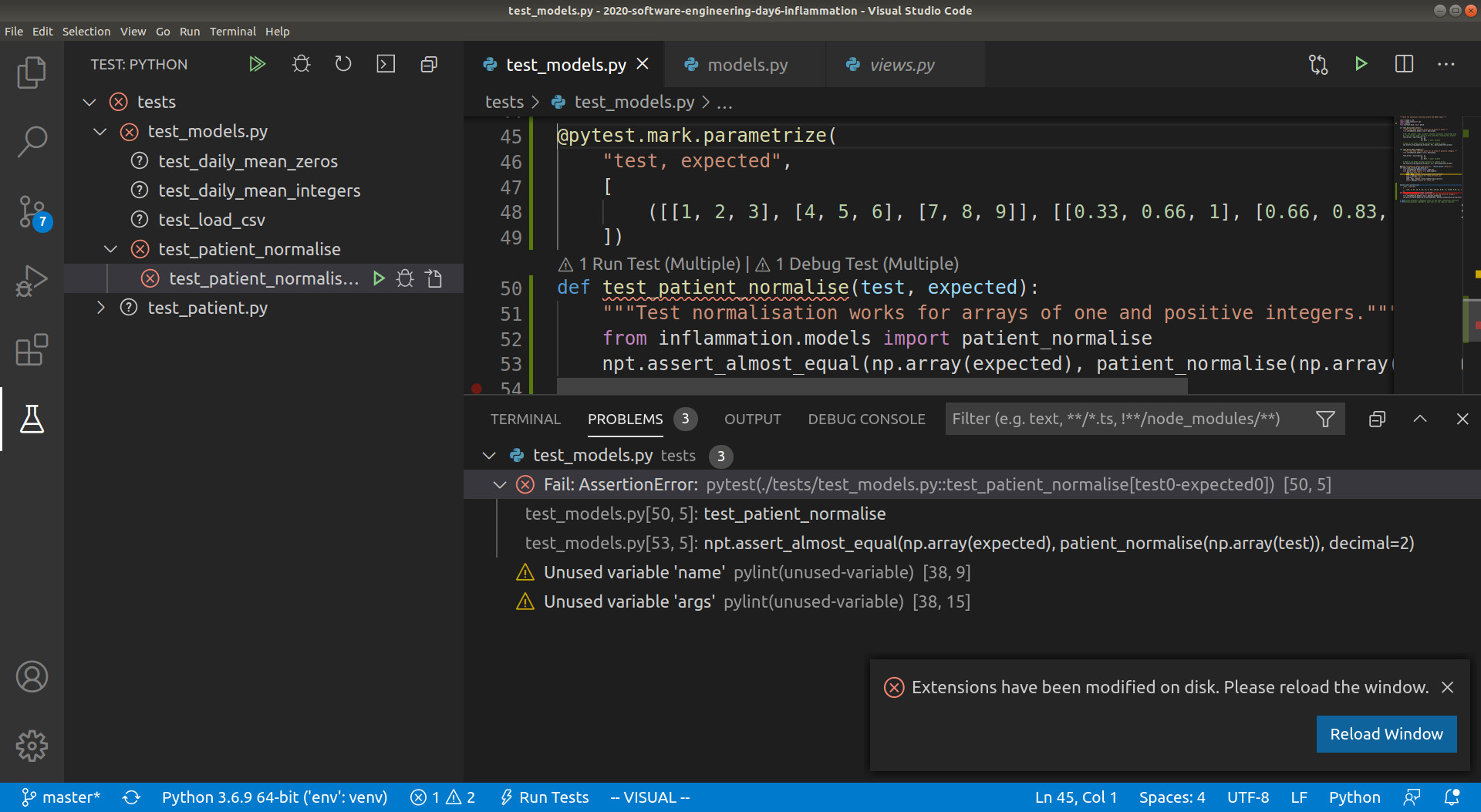
Click on the “run” button next to test_patient_normalise, and you will be able to see
that VSCode runs the function, and the same AssertionError that we say before.
Now we want to use the debugger to investigate what is happening inside the
patient_normalise function. To do this we will add a breakpoint in the code.
Navigate to the models.py file and move your mouse to the return statement of the
patient_normalise function. Click to the left of the line number for that line and a
small red dot will appear, indicating that you have placed a breakpoint on that line.
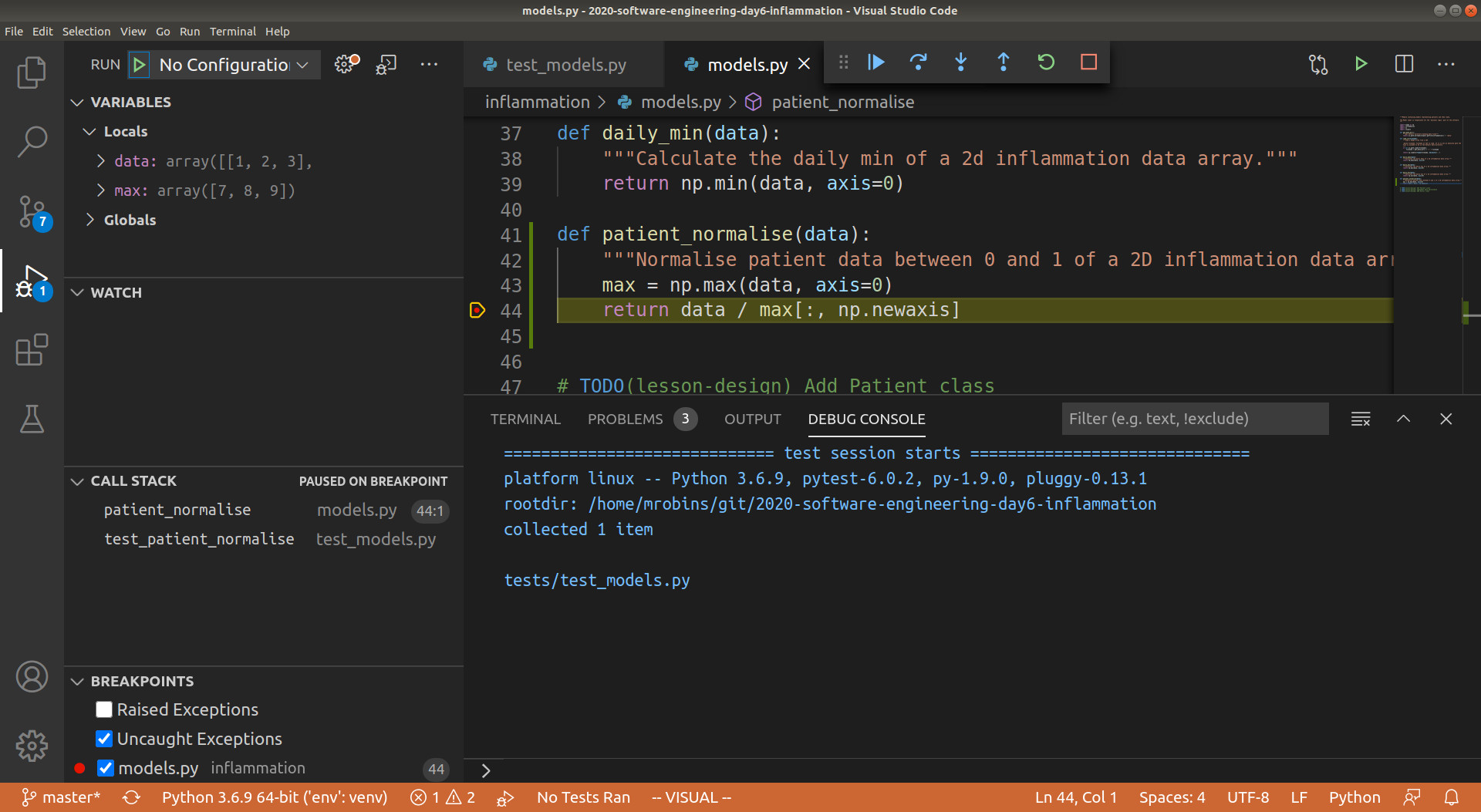
Now if you debug test_patient_normalise, you will notice that execution will be paused
at the return statement of patient_normalise, and we can investigate the exact state
of the program as it is executing this line of code. Navigate to the Run view, and you
will be able to see the local and global variables currently in memory, the call stack
(i.e. what functions are currently running), and the current list of breakpoints. In the
local variables section you will be able to see the data array that is input to the
patient_normalise function, as well as the max local array that was created to hold
the maximum inflammation values for each patient. See below for a screenshot.
In the Watch section of the Run view you can write any expression you want the debugger
to calculate, this is useful if you want to view a particular combination of variables,
or perhaps a single element or slice of an array. Try putting in the expression max[:,
np.newaxis] into the Watch section, and you will be able to see the column vector that
we are dividing data by in the return line of the function. You can also open the
Debug Console and type in max[:, np.newaxis] to see the same result.
Looking at the max variable, we can see that something looks wrong, as the maximum
values for each patient do not correspond to the data array. Recall that the input
data array we are using for the function is
[[1, 2, 3], [4, 5, 6], [7, 8, 9]]
so the maximum inflammation for each patient should be [3, 6, 9], whereas the debugger
shows [7, 8, 9]. You can see that the latter corresponds exactly to the last column of
data, and we can immediately conclude that we took the maximum along the wrong axis of
data. So to fix the function we can change axis=0 in the first line to axis=1.
With this fix in place, running the tests again will result in a passing test, and a
nice green tick next to the test in the VSCode IDE.
Corner or Edge Cases
The test case that we have currently written for patient_normalise is parameterised
with a fairly standard data array. However, when writing your test cases, it is
important to consider parametrising them by unusual or extrema values, in order to test
all the edge or corner cases that your code could be exposed to in practice. Generally
speaking, it is at these extrema cases that you will find your code failing, so it
beneficial to test them beforehand.
What is considered an “edge case” for any given component depends on what that component
is meant to do. In the case of patient_normalise the goal of the function is to
normalise a numeric array of numbers. For numerical values the extrema cases could be
zeros, very large or small values, not-a-number (NaN), or infinity values. Since we are
specifically considering an array of values, an edge case could be that all the
numbers of the array are equal.
For all the given edge cases you might come up with, you should also consider their
likelihood of occurrence, it is often too much effort to exhaustively test a given
function against every possible input, so you should prioritise edge cases that are
likely to occur. For our patient_normalise function, some common edge cases might be
the occurrence of zeros, and the case where all the values of the array are the same.
When you are considering edge cases to test for, try also to think about what might
break your code. For patient_normalise we can see that there is a division by the
maximum inflammation value for each patient, so this will clearly break if we are
dividing by zero here, resulting in NaN values in the normalised array.
With all this in mind, lets add a few edge cases to our parametrisation of
test_patient_normalise. We will add two extra tests, corresponding to an input array
of all 0, and an input array of all 1.
@pytest.mark.parametrize(
"test, expected",
[
([[0, 0, 0], [0, 0, 0], [0, 0, 0]], [[0, 0, 0], [0, 0, 0], [0, 0, 0]]),
([[1, 1, 1], [1, 1, 1], [1, 1, 1]], [[1, 1, 1], [1, 1, 1], [1, 1, 1]]),
([[1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0.33, 0.66, 1], [0.66, 0.83, 1], [0.77, 0.88, 1]]),
])
Running the tests now results in the following assertion error, due to the division by zero as we predicted.
E AssertionError:
E Arrays are not almost equal to 2 decimals
E
E x and y nan location mismatch:
E x: array([[0, 0, 0],
E [0, 0, 0],
E [0, 0, 0]])
E y: array([[nan, nan, nan],
E [nan, nan, nan],
E [nan, nan, nan]])
env/lib/python3.6/site-packages/numpy/testing/_private/utils.py:740: AssertionError
Helpfully, you will also notice that Numpy also provides a run-time warning for the divide by zero, reproduced below
RuntimeWarning: invalid value encountered in true_divide
return data / max[:, np.newaxis]
How can we fix this? Luckily there is a Numpy function that is useful here,
np.isnan(), which
we can use to replace all the NaN’s with our desired result, which is 0. We can also
silence the run-time warning using
np.errstate.
Exploring tests for edge cases
Fix the failing
test_patient_normalisetest, and think of some more suitable edge cases to test ourpatient_normalise()function and add them to the parametrised tests. After you have finished remember to commit your changes.Possible Solution
... def patient_normalise(data): """ Normalise patient data between 0 and 1 of a 2D inflammation data array. Any NaN values are ignored, and normalised to 0 Any negative values are clipped to 0 """ max = np.nanmax(data, axis=1) with np.errstate(invalid='ignore', divide='ignore'): normalised = data / max[:, np.newaxis] normalised[np.isnan(normalised)] = 0 normalised[normalised < 0] = 0 return normalised ... @pytest.mark.parametrize( "test, expected", [ ( [[0, 0, 0], [0, 0, 0], [0, 0, 0]], [[0, 0, 0], [0, 0, 0], [0, 0, 0]], ), ( [[1, 1, 1], [1, 1, 1], [1, 1, 1]], [[1, 1, 1], [1, 1, 1], [1, 1, 1]], ), ( [[float('nan'), 1, 1], [1, 1, 1], [1, 1, 1]], [[0, 1, 1], [1, 1, 1], [1, 1, 1]], ), ( [[1, 2, 3], [4, 5, float('nan')], [7, 8, 9]], [[0.33, 0.66, 1], [0.8, 1, 0], [0.77, 0.88, 1]], ), ( [[-1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0, 0.66, 1], [0.66, 0.83, 1], [0.77, 0.88, 1]], ), ( [[1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0.33, 0.66, 1], [0.66, 0.83, 1], [0.77, 0.88, 1]], ) ]) def test_patient_normalise(test, expected): """Test normalisation works for arrays of one and positive integers.""" from inflammation.models import patient_normalise npt.assert_almost_equal(np.array(expected), patient_normalise(np.array(test)), decimal=2) ...You could also test, and handle, the case of a whole row of NaNs…
Defensive programming to avoid potential errors
In the previous section, we have made a few design choices for our patient_normalise
function. The first was that we are implicitly converting any NaN and negative values to
0, the second is that normalising a constant 0 array of inflammation will result in an
identical array of 0’s. The third is that we don’t want to warn the user in any of these
situations. This could be handled differently, we might decide that we don’t want to
silently make these changes to the data, but instead to explicitly check that the input
data satisfies a given set of assumptions (e.g. no negative values), and raise an error
if this is not the case. Then we can proceed with the normalisation, confident that our
normalisation function will work correctly.
Checking valid input to a function via preconditions is one of the simplest forms of
defensive programming. These preconditions are checked at the beginning of the
function to make sure that all assumptions are satisfied. These assumptions are often
based on the value of the arguments, like we have already discussed. However, in a
dynamic language like Python one of the more common preconditions is to check that the
arguments of a function are of the correct type. Currently there is nothing stopping
someone from calling patient_normalise with a string, a dictionary, or another object
that is not an ndarray.
As an example, let’s change the behaviour of the patient_normalise function to raise
an error on negative inflammation values. We can add a precondition check to the
beginning of our function like so
...
if np.any(data < 0):
raise ValueError('inflammation values should be non-negative')
...
We can then modify our test function to check that the function raises the correct
exception, a ValueError. The
ValueError exception
is part of the standard library and is used to indicate that the function received an
argument of the right type, but an inappropriate value.
@pytest.mark.parametrize(
"test, expected, raises",
[
... # other test cases here, with None for raises
(
[[-1, 2, 3], [4, 5, 6], [7, 8, 9]],
[[0, 0.66, 1], [0.66, 0.83, 1], [0.77, 0.88, 1]],
ValueError,
),
(
[[1, 2, 3], [4, 5, 6], [7, 8, 9]],
[[0.33, 0.66, 1], [0.66, 0.83, 1], [0.77, 0.88, 1]],
None,
),
])
def test_patient_normalise(test, expected, expected_error):
"""Test normalisation works for arrays of one and positive integers."""
from inflammation.models import patient_normalise
if expected_error:
with pytest.raises(expected_error):
npt.assert_almost_equal(np.array(expected), patient_normalise(np.array(test)), decimal=2)
else:
npt.assert_almost_equal(np.array(expected), patient_normalise(np.array(test)), decimal=2)
Add precondition checking correct type and shape of data
We are not currently checking that the
dataargument totest_patient_normaliseis of a valid type. Add one precondition to check that data is anndarrayobject, and another to check that it is of the correct shape. Add corresponding tests to check that the function raises the correct exception. You will probably find the Python functionisinstanceuseful here, as well as the Python exceptionTypeError. Once you are done, commit your new files, and push the new commits to your remote repository on GitHubSolution
... def patient_normalise(data): """ Normalise patient data between 0 and 1 of a 2D inflammation data array. Any NaN values are ignored, and normalised to 0 :param data: 2d array of inflammation data :type data: ndarray """ if not isinstance(data, np.ndarray): raise TypeError('data input should be ndarray') if len(data.shape) != 2: raise ValueError('inflammation array should be 2-dimensional') if np.any(data < 0): raise ValueError('inflammation values should be non-negative') max = np.nanmax(data, axis=1) with np.errstate(invalid='ignore', divide='ignore'): normalised = data / max[:, np.newaxis] normalised[np.isnan(normalised)] = 0 return normalised ... @pytest.mark.parametrize( "test, expected, raises", [ ... ( 'hello', None, TypeError, ), ( 3, None, TypeError, ), ( [[1, 2, 3], [4, 5, 6], [7, 8, 9]], [[0.33, 0.66, 1], [0.66, 0.83, 1], [0.77, 0.88, 1]], None, ) ]) def test_patient_normalise(test, expected, raises): """Test normalisation works for arrays of one and positive integers.""" from inflammation.models import patient_normalise if isinstance(test, list): test = np.array(test) if raises: with pytest.raises(raises): npt.assert_almost_equal(np.array(expected), patient_normalise(test), decimal=2) else: npt.assert_almost_equal(np.array(expected), patient_normalise(test), decimal=2) ...
Don’t take it too far and try to code preconditions for every conceivable eventuality.
You must always strike a balance between making sure you secure your function against
incorrect use, and writing an overly complicated and expensive function that handles
cases that could never possibly occur. For example, it would be sensible to validate the
shape of your inflammation data array when it is actually read from the csv file (in
load_csv), and therefore there is no reason to test this again in patient_normalise.
You can always also neglect to add explicit preconditions in your code, but instead
state the assumptions and limitations of your code for others in the docstring, trusting
that they will use the function correctly. This approach is useful when explicitly
checking the precondition would be too costly to execute.
Key Points
Unit testing is good to show us what doesn’t work, but does not help us locate problems.
We can use a debugger to help us locate problems in our program.
A debugger allows us to pause a program and examine it’s state by adding breakpoints to lines in code.
We can use preconditions to ensure correct behaviour in our programs.
We must ensure our unit tests cater for edge and corner cases sufficiently.